PALM Create Static Driver (palm_csd)
In order to create comprehensive drivers for PALM runs in urban environments, PALM comes shipped with the tool palm_csd. This python3-based script was developed in the course of the MOSAIK project and thus is, in principal, only working for the democities Berlin, Hamburg, and Stuttgart. However, with some effort it is possible to adapt the script for other cities as well. Note that in the mid-term, palm_csd will be replaced by a more generic and universal tool based with a graphical user interface as front end.
Usage
Execution
In order to create a static driver, go to the working directory (usually ~/palm/current_version), prepare a palm_csd configuration file (see below, an exemplary file can be found here). After set-up of the configuration file, execute palm_csd as follows:
palm_csd .csd.config.default
The static driver will be written to the directory specified in the configuration file. During compilation of the driver, palm_csd will print some more or less useful information to screen.
Configuration file
This section describes how to set-up a configuration file for creating a static driver for PALM based on pre-processed NetCDF data. In the following we will use data for Berlin, which is available via Open Access. The configuration file uses the well-known format of INI files, which are processed by the ConfigParser? package in python. It consists of the following sections:
[global]
A set of global attributes can be defined that will be passed to the static driver file. In details, the following attributes can be set (see also static driver description:
| Attribute | Data type | Default value | Description |
|---|---|---|---|
| author | string | "" | Author of the static driver. Use the format: name, email |
| contact_person | string | "" | Contact person, format as for author |
| acronym | string | "" | Institutional acronym (formatting rules apply to static drivers used in the MOSAIK project) |
| comment | string | "" | Arbitrary text |
| data_content | string | "" | Arbitrary text |
| dependencies | string | "" | Arbitrary text |
| keywords | string | "" | Arbitrary keywords |
| source | string | "" | List of data sources used to generate the driver |
| campaign | string | "" | Information on measurement capaign (if applicable) |
| location | string | "" | Geo-location of the static driver content (if applicable) |
| site | string | "" | Site description od the static driver content (if applicable) |
| institution | string | "" | Institution of the driver creator |
| palm_version | float | "" | PALM version for which the driver was generated (for comptatibility checks) |
| rotation_angle | float | 0.0 | Clockwise angle of rotation in degrees between North positive y axis and the y axis in the data, e.g. 0.0. This value overwrites the namelist parameter rotation_angle |
| references | string | "" | Arbitrary text |
Example:
author = Bjoern Maronga, maronga@meteo.uni-hannover.de contact_person = Bjoern Maronga, maronga@meteo.uni-hannover.de acronym = LUHimuk comment = created with palm_csd data_content dependencies keywords source campaign location = Berlin site = Berlin Mitte institution = Institute of Meteorology and Climatology, Leibniz University Hannover palm_version = 6.0 rotation_angle = 0.0 references
Note that most of the global attributes have no effect on the PALM simulations, except rotation_angle.
[settings]
This section describes global parameters used to create the static driver.
| Variable | Data type | Default value | Description |
|---|---|---|---|
| bridge_width | float | 3.0 | In case that the simulation domain contains bridges, this parameter (in m) defines the vertical thickness of all bridge elements in the domain. Note that bridges require LOD2 building information (i.e. buildings_3d) |
| lai_roof_extensive | float | 0.8 | Leaf are index for green roofs with extensive vegetation, defined by setting the appropriate building_pars field. The value is assigned to all extensive green roofs in the model domain |
| lai_roof_intensive | float | 2.5 | Leaf are index for green roofs with intensive vegetation, defined by setting the appropriate building_pars field. The value is assigned to all intensive green roofs in the model domain |
| lai_high_vegetation_default | float | 6.0 | Default leaf area index for (high) vegetation used to generate the 3D leaf area density field. This value is used for all pixels for which no other leaf area density is available (i.e. to fill missing data) |
| lai_low_vegetation_default | float | 1.0 | Default leaf area index for (low) vegetation used to fill data gaps in the leaf area index distribution. This parameter only will a LOD2 leaf area index for parameterized vegetation via vegetation_type, i.e., through the vegetation_pars field |
| lai_tree_lower_threshold | float | 0.0 | Lower threshold of LAI for trees. Trees with LAI < lai_tree_lower_threshold are either removed or considered to have LAI=lai_tree_lower_threshold, depending on the setting remove_low_lai_tree. |
| lai_alpha | float | 5.0 | Parameter for reconstruction of vertical LAD profiles based on tree shape parameters (alpha, beta) and the integral leaf area index after Markkanen et al. (2003). This scheme is used for vegetation patches (parks, forests), where the canopy can be considered to be pseudo-1D and for which usually no information on individual trees is available. |
| lai_beta | float | 3.0 | Parameter for reconstruction of vertical LAD profiles based on tree shape parameters (alpha, beta) and the integral leaf area index after Markkanen et al. (2003). This scheme is used for vegetation patches (parks, forests), where the canopy can be considered to be pseudo-1D and for which usually no information on individual trees is available. |
| patch_height_default | float | 10.0 | Default patch height (in m), which is used in the canopy generator to process canopy patches (parks, forests) for which data for individual trees is usually lacking. This parameter comes into affect for data gaps where no other vegetation height is available |
| season | string | summer | As palm_csd can work with different sets of input data regarding leaf area index, this switch parameter can be set to either summer or winter to select the most suitable leaf area index input file to account for differences in leaf amount. Data for summer is usually from August (fully leaved), while data for winter is usually from April. |
| vegetation_type_below_trees | integer | 3 | If trees are added to the static driver, the vegetation type below the tree volumes is changed to this value. |
Example:
bridge_width = 3.0 debug_mode = False lai_roof_extensive = 3.0 lai_roof_intensive = 1.5 lai_high_vegetation_default = 5.0 lai_low_vegetation_default = 1.0 lai_alpha = 5.0 lai_beta = 3.0 patch_height_default = 10.0 season = summer
[output]
This section describes the location for the static driver output.
| Variable | Data type | Default value | Description |
|---|---|---|---|
| path | string | "" | Directory where the output file shall be stored. Note that the static driver can - depending on model domain size - be quite large (in the order of several GB). |
| file_out* | string | Output file name. The final output will be stored under path/file_out_domain, where domain will be "root" for the parent (root) domain, and "N01", "N02", etc., for child domains N01, N02, etc., respectively | |
| version | integer | "" | User-specific setting to track updates of a static driver. This value will be added as global attribute to the static driver |
\* This parameter is mandatory
Example:
path = /ldata2/MOSAIK/ file_out = winter_iop1_test version = 1
[input_01] - [input_XX]
The configuration file can include several sets of input data for different grid spacing. For each set of input data, an individual section must be provided and numbered accordingly (i.e., [input_01], [input_02], etc.).
| Variable | Data type | Description |
|---|---|---|
| path | string | Directory where the NetCDF input files reside |
| pixel_size* | float | Horizontal grid spacing (m) of a surface pixel of size dx*dy where dx = dy in the input data |
| file_x* | string | UTM x-coordinates for the simulation domain (m) |
| file_y* | string | UTM y-coordinates for the simulation domain (m) |
| file_x_UTM* | string | UTM x-coordinates for the simulation domain (m) |
| file_y_UTM* | string | UTM y-coordinates for the simulation domain (m) |
| file_lat* | string | Latitude (degrees N) for the simulation domain |
| file_lon* | string | Longitude (degrees E) for the simulation domain |
| file_zt* | string | Terrain height (m) |
| file_buildings_2d* | string | 2D Building height (m) |
| file_building_id* | string | Building ids |
| file_building_type* | string | Building type distribution |
| file_bridges_2d* | string | 2D Map of bridge height (m) |
| file_bridges_id* | string | Bridge ids |
| file_lai | string | Leaf area index |
| file_vegetation_type* | string | Vegetation type distribution |
| file_vegetation_height* | string | Vegetation height (m) |
| file_pavement_type* | string | Pavement type distribution |
| file_water_type* | string | Water type distribution |
| file_street_type* | string | Street type distribution (used for parameterized chemistry emissions and multi-agent model) |
| file_street_crossings* | string | Street crossings (used for multi-agent model) |
| file_tree_height | string | Tree height (m) for street trees. For each tree only one value can be given at the center of the tree location |
| file_tree_crown_diameter | string | Tree crown diameter (m). For each tree only one value can be given at the center of the tree location |
| file_tree_trunk_diameter | string | Trunk diameter at breast height (m). For each tree only one value can be given at the center of the tree location |
| file_tree_type* | string | Tree type according to the canopy generator tree inventory. For each tree only one value can be given at the center of the tree location |
| file_patch_height* | string | 2D distribution of the vegetation canopy height |
| file_patch_type | string | 2D distribution of the vegetation type of vegetation patches |
| file_vegetation_on_roofs | string | 2D distribution of green roofs. Values can be 0.0-1.0. Intensive vegetation is considered for values >= 0.5, while extensive vegetation is assumed for values > 0.5 |
\* This parameter is mandatory
Example:
path = /ldata2/MOSAIK/Berlin_static_driver_data pixel_size = 15.0 file_x = Berlin_CoordinatesUTM_y_15m_DLR.nc file_y = Berlin_CoordinatesUTM_x_15m_DLR.nc file_lat = Berlin_CoordinatesLatLon_y_15m_DLR.nc file_lon = Berlin_CoordinatesLatLon_x_15m_DLR.nc file_zt = Berlin_terrain_height_15m_DLR.nc file_buildings_2d = Berlin_building_height_15m_DLR.nc file_building_id = Berlin_building_id_15m_DLR.nc file_building_type = Berlin_building_type_15m_DLR.nc file_bridges_2d = Berlin_bridges_height_15m_DLR.nc file_bridges_id = Berlin_bridges_id_15m_DLR.nc file_lai = Berlin_leaf_area_index_15m_DLR_WANG_summer.nc file_vegetation_type = Berlin_vegetation_type_15m_DLR.nc file_vegetation_height = Berlin_vegetation_patch_height_15m_DLR.nc file_pavement_type = Berlin_pavement_type_15m_DLR.nc file_water_type = Berlin_water_type_15m_DLR.nc file_soil_type = Berlin_soil_type_15m_DLR.nc file_street_type = Berlin_street_type_15m_DLR.nc file_street_crossings = Berlin_street_crossings_15m_DLR.nc file_tree_height = Berlin_trees_height_clean_15m.nc file_tree_crown_diameter = Berlin_tree_crown_15m_DLR.nc file_tree_trunk_diameter = Berlin_trees_trunk_clean_15m.nc file_tree_type = Berlin_trees_type_15m_DLR.nc file_patch_height = Berlin_vegetation_patch_height_15m_DLR.nc file_vegetation_on_roofs = Berlin_vegetation_on_roofs_15m_DLR.nc
[domain_root] - [domain_XXX]
This section contains settings for each model domain for the PALM run. The section for the root domain domain must be named [domain_root]. In case of non-nested runs, this is the default model domain. In case of a nested run, the sections for the non-root domains must be named [domain_N01], [domain_N02], etc. as it is done in the PALM parameter file.
| Variable | Data type | Default value | Description |
|---|---|---|---|
| pixel_size* | float | Grid spacing in x and y / pixel size (m) | |
| origin_x* | float | x-origin of the model domain (m) with respect to the input data UTM coordinates | |
| origin_y* | float | y-origin of the model domain (m) with respect to the input data UTM coordinates | |
| nx* | float | Number of grid points in x-direction (equals the nx setting in the PALM parameter file | |
| ny* | float | Number of grid points in y-direction (equals the ny setting in the PALM parameter file | |
| dz* | float | Vertical grid spacing in PALM (m). This parameter is needed when buildings_3d}, {{{street_trees, canopy_patches, interpolate_terrain, or use_palm_z_axis is used | |
| buildings_3d | logical | False | Use 3D buildings via the buildings_3d array to buildings instead of buildings_2d. This parameter must be true if bridges are present in the simulation domain. Note that the processing of 3D buildings by palm_csd is slower than 2D buildings |
| allow_high_vegetation | logical | False | If set to True, it is allowed to have high vegetation classes according in the vegetation_type distribution. Note that this can involve very large roughness lengths > 0.5 m. If the vertical grid spacings is close or smaller than this threshold the PALM run will crash and/or does not provide meaningful results. It is generally recommended to set this parameter to False whenever the grid spacing in small enough to resolve canopy patches be 2 or more vertical grid levels. If set to False pixels where a high vegetation type was prescribed will be converted into a 3D leaf area density canopy using the canopy generator |
| generate_vegetation_patches | logical | True | If set to True the embedded canopy generator will convert all surface pixels that contain high vegetation into a 3D leaf area density distribution. This applies to pixels where vegetation_type is set to a high vegetation type, or where the vegetation height field suggests high vegetation. Note that only pixels with heights > 2*dz are converted, while all other pixels will be parameterized via the vegetation_type field |
| use_palm_z_axis | logical | False | If set to True, the static driver will raster the input data on the z-grid of PALM for output. Note that PALM will convert continuous static driver data itself on its grid and apply additional filtering procedures. It is thus recommended to set this parameter to False unless interpolate_terrain = True in nested set-ups. |
| interpolate_terrain | logical | False | If set to True, the terrain height is interpolated and blended over between parent and child domains in order to avoid severe steps in terrain height due to different grid spacings between parent and child |
| domain_parent | string | Name of the parent domain of the current domain. If the current domain is the root domain, leave this parameter empty. | |
| vegetation_on_roofs | logical | True | If set to True, allow green roofs |
| street_trees | logical | True | If set to true, information on individual street trees will be used to generate a 3D leaf area density and basal area density distribution for each tree. In contrast to vegetation patches, where a closed canopy is assumed and information is only distributed vertically for each pixel, street trees have a 3D shape that is mapped on the simulation domain |
| overhanging_trees | logical | True | If set to False, no LAD volumes of trees are generated above surfaces without a vegetation type. |
| remove_low_lai_tree | logical | False | If set to True, all trees with an LAI<lai_tree_lower_threshold are removed from the dataset. If set to False, those trees are considered with LAI=lai_tree_lower_threshold. |
\* This parameter is mandatory
Example:
pixel_size = 15.0 origin_x = 19605 origin_y = 20895 nx = 199 ny = 199 buildings_3d = False dz = 15.0 allow_high_vegetation = True generate_vegetation_patches = True use_palm_z_axis = False interpolate_terrain = False domain_parent vegetation_on_roofs = True street_trees = True
Required input data
At the moment it is required to provide all input files as listed under [input_XX]. For a given pixel size (i.e. horizontal grid spacing), only one set of input files can be provided. All input data must be two-dimensions (y,x). Currently, palm_csd expects all variables in the respective input files to be named 'Band1'. It is planned to implement a more flexible interface in the near future.
Technical documentation
Under construction.
Overview
Processing of nested domains
Canopy generator
Single trees
Tree database
Default values for trees if individual parameters are not provided. Default data is derived as mean values from the tree database for Berlin, Germany.
| Index | |=Species | Shape | Crown height/width ratio(*) | Crown diameter (m) | Height (m) | LAI summer(*) | LAI winter(*) | Height of maximum LAD (m)(*) | LAD/BAD ratio(*) | DBH (m) |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Default | 1.0 | 1.0 | 4.0 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.35 |
| 1 | Abies | 3.0 | 1.0 | 4.0 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 2 | Acer | 1.0 | 1.0 | 7.0 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 3 | Aesculus | 1.0 | 1.0 | 7.0 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.00 |
| 4 | Ailanthus | 1.0 | 1.0 | 8.5 | 13.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.30 |
| 5 | Alnus | 3.0 | 1.0 | 6.0 | 16.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.20 |
| 6 | Amelanchier | 1.0 | 1.0 | 3.0 | 4.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.20 |
| 7 | Betula | 1.0 | 1.0 | 6.0 | 14.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.30 |
| 8 | Buxus | 1.0 | 1.0 | 4.0 | 4.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 9 | Calocedrus | 3.0 | 1.0 | 5.0 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 10 | Caragana | 1.0 | 1.0 | 3.5 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 11 | Carpinus | 1.0 | 1.0 | 6.0 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 12 | Carya | 1.0 | 1.0 | 5.0 | 17.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 13 | Castanea | 1.0 | 1.0 | 4.5 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 14 | Catalpa | 1.0 | 1.0 | 5.5 | 6.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 15 | Cedrus | 1.0 | 1.0 | 8.0 | 13.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 16 | Celtis | 1.0 | 1.0 | 6.0 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 17 | Cercidiphyllum | 1.0 | 1.0 | 3.0 | 6.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 18 | Cercis | 1.0 | 1.0 | 2.5 | 7.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 19 | Chamaecyparis | 5.0 | 1.0 | 3.5 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 20 | Cladrastis | 1.0 | 1.0 | 5.0 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 21 | Cornus | 1.0 | 1.0 | 4.5 | 6.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.20 |
| 22 | Corylus | 1.0 | 1.0 | 5.0 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
| 23 | Cotinus | 1.0 | 1.0 | 4.0 | 4.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 24 | Crataegus | 3.0 | 1.0 | 3.5 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.40 |
| 25 | Cryptomeria | 3.0 | 1.0 | 5.0 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 26 | Cupressocyparis | 3.0 | 1.0 | 3.0 | 8.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
| 27 | Cupressus | 3.0 | 1.0 | 5.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
| 28 | Cydonia | 1.0 | 1.0 | 2.0 | 3.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 29 | Davidia | 1.0 | 1.0 | 10.0 | 14.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
| 30 | Elaeagnus | 1.0 | 1.0 | 6.5 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.20 |
| 31 | Euodia | 1.0 | 1.0 | 4.5 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 32 | Euonymus | 1.0 | 1.0 | 4.5 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 33 | Fagus | 1.0 | 1.0 | 10.0 | 12.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 34 | Fraxinus | 1.0 | 1.0 | 5.5 | 10.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.60 |
| 35 | Ginkgo | 3.0 | 1.0 | 4.0 | 8.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 36 | Gleditsia | 1.0 | 1.0 | 6.5 | 10.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 37 | Gymnocladus | 1.0 | 1.0 | 5.5 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 38 | Hippophae | 1.0 | 1.0 | 9.5 | 8.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 39 | Ilex | 1.0 | 1.0 | 4.0 | 7.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 40 | Juglans | 1.0 | 1.0 | 7.0 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 41 | Juniperus | 5.0 | 1.0 | 3.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 42 | Koelreuteria | 1.0 | 1.0 | 3.5 | 5.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 43 | Laburnum | 1.0 | 1.0 | 3.0 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 44 | Larix | 3.0 | 1.0 | 7.0 | 16.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 45 | Ligustrum | 1.0 | 1.0 | 3.0 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 46 | Liquidambar | 3.0 | 1.0 | 3.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.30 |
| 47 | Liriodendron | 3.0 | 1.0 | 4.5 | 9.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 48 | Lonicera | 1.0 | 1.0 | 7.0 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 49 | Magnolia | 1.0 | 1.0 | 3.0 | 5.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 50 | Malus | 1.0 | 1.0 | 4.5 | 5.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.30 |
| 51 | Metasequoia | 5.0 | 1.0 | 4.5 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 52 | Morus | 1.0 | 1.0 | 7.5 | 11.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.00 |
| 53 | Ostrya | 1.0 | 1.0 | 2.0 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.00 |
| 54 | Parrotia | 1.0 | 1.0 | 7.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.30 |
| 55 | Paulownia | 1.0 | 1.0 | 4.0 | 8.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
| 56 | Phellodendron | 1.0 | 1.0 | 13.5 | 13.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 57 | Picea | 3.0 | 1.0 | 3.0 | 13.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.90 |
| 58 | Pinus | 3.0 | 1.0 | 6.0 | 16.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 59 | Platanus | 1.0 | 1.0 | 10.0 | 14.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 60 | Populus | 1.0 | 1.0 | 9.0 | 20.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.40 |
| 61 | Prunus | 1.0 | 1.0 | 5.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.60 |
| 62 | Pseudotsuga | 3.0 | 1.0 | 6.0 | 17.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 63 | Ptelea | 1.0 | 1.0 | 5.0 | 4.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 64 | Pterocaria | 1.0 | 1.0 | 10.0 | 12.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 65 | Pterocarya | 1.0 | 1.0 | 11.5 | 14.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.60 |
| 66 | Pyrus | 3.0 | 1.0 | 3.0 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.80 |
| 67 | Quercus | 1.0 | 1.0 | 8.0 | 14.0 | 3.1 | 0.1 | 0.6 | 0.025 | 0.40 |
| 68 | Rhamnus | 1.0 | 1.0 | 4.5 | 4.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.30 |
| 69 | Rhus | 1.0 | 1.0 | 7.0 | 5.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 70 | Robinia | 1.0 | 1.0 | 4.5 | 13.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 71 | Salix | 1.0 | 1.0 | 7.0 | 14.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 72 | Sambucus | 1.0 | 1.0 | 8.0 | 6.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.40 |
| 73 | Sasa | 1.0 | 1.0 | 10.0 | 25.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 74 | Sequoiadendron | 5.0 | 1.0 | 5.5 | 10.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.60 |
| 75 | Sophora | 1.0 | 1.0 | 7.5 | 10.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.40 |
| 76 | Sorbus | 1.0 | 1.0 | 4.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 77 | Syringa | 1.0 | 1.0 | 4.5 | 5.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 78 | Tamarix | 1.0 | 1.0 | 6.0 | 7.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.50 |
| 79 | Taxodium | 5.0 | 1.0 | 6.0 | 16.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.60 |
| 80 | Taxus | 2.0 | 1.0 | 5.0 | 7.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.50 |
| 81 | Thuja | 3.0 | 1.0 | 3.5 | 9.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 82 | Tilia | 3.0 | 1.0 | 7.0 | 12.5 | 3.0 | 0.8 | 0.6 | 0.025 | 0.70 |
| 83 | Tsuga | 3.0 | 1.0 | 6.0 | 10.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.10 |
| 84 | Ulmus | 1.0 | 1.0 | 7.5 | 14.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.80 |
| 85 | Zelkova | 1.0 | 1.0 | 4.0 | 5.5 | 3.0 | 0.8 | 0.6 | 0.025 | 1.20 |
| 86 | Zenobia | 1.0 | 1.0 | 5.0 | 5.0 | 3.0 | 0.8 | 0.6 | 0.025 | 0.40 |
(*) Preliminary parameter.
Vegetation canopies
Best practices
The following example is a best practice setting for a high-resolution (1 m grid spacing) non-nested run in which most of the vegetation can be resolved via a 3D leaf area density distribution:
[domain_root] domain_parent pixel_size = 1.0 origin_x = ... origin_y = ... nx = ... ny = ... dz = ... allow_high_vegetation = False buildings_3d = True generate_vegetation_patches = True use_palm_z_axis= False interpolate_terrain = False vegetation_on_roofs = True street_trees = True
For a nested run, the following settings should work nicely to avoid terrain height issues:
[domain_root] domain_parent pixel_size = 15.0 dz = 15.0 origin_x = ... origin_y = ... nx = ... ny = ... buildings_3d = False allow_high_vegetation = True generate_vegetation_patches = True use_palm_z_axis= False interpolate_terrain = False vegetation_on_roofs = False street_trees = True [domain_N02] domain_parent = root pixel_size = 1.0 dz = 1.0 origin_x = ... origin_y = ... nx = ... ny = ... buildings_3d = True allow_high_vegetation = False generate_vegetation_patches = True use_palm_z_axis= True interpolate_terrain = True vegetation_on_roofs = True street_trees = True
Example static driver file
An example static driver file can be downloaded here (built for PALM r4311).
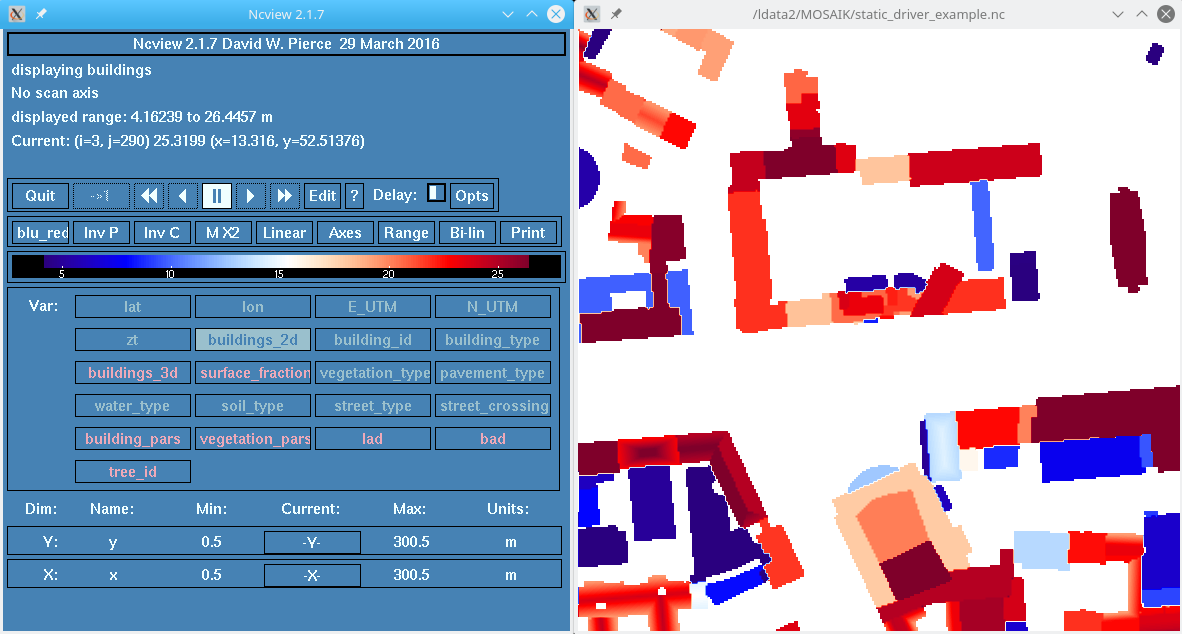 Snapshot of building heights in example static driver file (using ncview) |  ncdump of the static driver file header |
Literature
- Heldens, W., Burmeister, C., Kanani-Sühring, F., Maronga, B., Pavlik, D., Sühring, M., Zeidler, J., and Esch, T.: Geospatial input data for the PALM model system 6.0: model requirements, data sources and processing, Geosci. Model Dev., 13, 5833–5873, https://doi.org/10.5194/gmd-13-5833-2020, 2020.
- Lalic, B. and Mihailovic, D. T., An Empirical Relation Describing Leaf-Area Density inside the Forest for Environmental Modeling. Journal of Applied Meteorology, vol. 43, no. 4, pp. 641–645, 2004. doi:10.1175/1520-0450(2004)043<0641:AERDLD>2.0.CO;2.
- Markkanen, T., Rannik, Ü., Marcolla, B. et al. Footprints and Fetches for Fluxes over Forest Canopies with Varying Structure and Density. Boundary-Layer Meteorology 106, 437–459 (2003). https://doi.org/10.1023/A:1021261606719
Attachments (5)
- screen_example.png (40.8 KB) - added by maronga 5 years ago.
- ncdump_screen.png (99.7 KB) - added by maronga 5 years ago.
- static_driver_example_dump.txt (9.1 KB) - added by maronga 5 years ago.
- csd.config.example (7.9 KB) - added by maronga 5 years ago.
- static_driver_example.nc (60.3 MB) - added by maronga 5 years ago.
