| Version 135 (modified by hoffmann, 13 years ago) (diff) |
|---|
PALM error messages ¶
In the following list the error messages reported by PALM are displayed. Each error message is identified by a unique message identifier which is composed of two letters and a number. The second column contains the error message which is reported in the job protocol (see here?). In addition, the third column includes further explanations of the error.
Identifier for messages in the PALM standard code | |
Identifier for messages in ''user_'' routines | |
Identifier for messages from the netCDF library |
| Message identifier | Error message | Explanation |
|---|---|---|
| PA0001 ¶ | "host" is not set. Please check that environment variable "localhost" is set before running PALM | |
| PA0002 ¶ | illegal coupling mode: ... | A atmosphere-ocean coupling is implemented in palm. The internal variable coupling_mode gives information about the current model setup. It is internally set to "atmosphere_to_ocean" (atmospheric part of a coupled run), "ocean_to_atmosphere (ocean part of a coupled run), "precursor_atmos" (a precursor atmosphere run), "precursor_ocean" (a precursor ocean run), or "uncoupled" (atmosphere or ocean run without any intended coupling). This error message suggests that the initialization of the coupling did fail. Possible reasons might be trying a coupling between two atmospere runs (or two ocean runs), or problems with the mrun script. |
| PA0003 ¶ | dt_coupling is not set but required for coupling mode "..." | This error appears if the temporal interval for the data exchange dt_coupling is not explicitely set in case of runs with coupled models (e.g. atmosphere - ocean). |
| PA0004 ¶ | coupling mode "...": dt_coupling = ... is not equal to dt_coupling_remote = ... | For coupled runs the temporal interval for the data exchange dt_coupling must be explicitely set and equal in both models. |
| PA0005 ¶ | coupling mode "...": dt_coupling <= 0.0 is not allowed and is reset to MAX(dt_max(A,O)) = ... | This error occurs if an invalid value for dt_coupling is used in the parameter file. dt_coupling must be > 0.0 and equal in both models. |
| PA0006 ¶ | coupling mode "...": restart_time = ... is not equal to restart_time_remote = ... | restart_time has not been set properly (equal) in the atmosphere and ocean parameter file. See also coupled runs. |
| PA0007 ¶ | coupling mode "...": dt_restart = ... is not equal to dt_restart_remote = ... | dt_restart has not been set properly (equal) in the atmosphere and ocean parameter file. See also coupled runs. |
| PA0008 ¶ | coupling mode "...": simulation_time_since_reference = ... is not equal to simulation_time_since_reference_remote = ... | The simulated time is stored internally in the variable simulation_time_since_reference. In coupled mode both times must be equal. This error might occur when precursor runs are used with different startup times and the coupling is initiated inappropiately. See coupled runs for a detailed description of the setup for precursor runs. |
| PA0009 ¶ | dx in atmosphere is not larger or equal than dx in ocean | Coupled runs allow to use different grid resolutions in atmosphere and ocean. It is, however, not possible to use a finer grid resolution in the atmosphere than in the ocean. |
| PA0010 ¶ | Domain size in x-direction is not equal in ocean and atmosphere | Coupled runs are only possible, if the horizontal model domain size is equal in both, atmosphere and ocean. Have a look at the settings for dx, dy, nx and .ny? to avoid this error. |
| PA0011 ¶ | dy in atmosphere is not larger or equal than dy in ocean | Coupled runs allow to use different grid resolutions in atmosphere and ocean. It is, however, not possible to use a finer grid resolution in the atmosphere than in the ocean. |
| PA0012 ¶ | Domain size in y-direction is not equal in ocean and atmosphere | Coupled runs are only possible, if the horizontal model domain size is equal in both, atmosphere and ocean. Have a look at the settings for dx, dy, nx and .ny? to avoid this error. |
| PA0013 ¶ | illegal value given for loop_optimization: "..." | |
| PA0014 ¶ | a non-flat topography does not allow ... | |
| PA0015 ¶ | ocean = .T. does not allow ... | |
| PA0016 ¶ | unknown solver for perturbation pressure: psolver = "..." | Siggi |
| PA0017 ¶ | psolver = "..." only works for a 1d domain-decomposition along x please do not set npey/=1 in the parameter file | Siggi |
| PA0018 ¶ | error number free to use | |
| PA0019 ¶ | psolver = "..." only works for a parallel environment | Siggi |
| PA0020 ¶ | unknown multigrid cycle: cycle_mg = "..." | Siggi |
| PA0021 ¶ | unknown fft-algorithm: fft_method = "..." | Siggi |
| PA0022 ¶ | unknown advection scheme: momentum_advec = "..." | Different momentum advection schemes are available in PALM. Please set momentum_advec to either 'ws-scheme' or 'pw-scheme' . |
| PA0023 ¶ | momentum_advec or scalar_advec = "..." is not allowed with timestep_scheme = "..." | 5th order advection scheme is conditional unstable in conjunction with 'euler' or 'runge-kutta-2' integration. Instead, use timestep_scheme='runge-kutta-3'. |
| PA0024 ¶ | unknown advection scheme: scalar_advec = "..." | Different scalar advection schemes are available in PALM. Please set scalar_advec to either 'ws-scheme', 'pw-scheme' or 'bc-scheme' . |
| PA0025 ¶ | use_upstream_for_tke set .TRUE. because use_sgs_for_particles = .TRUE. | |
| PA0026 ¶ | advection_scheme scalar_advec = "bc-scheme" not implemented for loop_optimization = "cache" | For the Bott-Chlond scheme loop_optimization = "vector" must to be set. |
| PA0027 ¶ | unknown timestep scheme: timestep_scheme = "..." | Different timestep schemes are available in PALM. Please set timestep_scheme to either 'runge-kutta-3' , 'runge-kutta-2' , or 'euler' . |
| PA0028 ¶ | error number free to use | |
| PA0029 ¶ | momentum advection scheme "..." does not work with timestep_scheme "..." | For timestep_scheme = 'runge-kutta-3' only momentum_advec = 'ws-scheme' or 'pw-scheme' lead to a numerically stable solution. |
| PA0030 ¶ | initializing_actions = "..." unkown or not allowed | For the initial run of a job chain one of the allowed values for initializing_actions must be set. For a restart run please set initializing_actions = 'read_restart_data' . |
| PA0031 ¶ | initializing_actions = "set_constant_profiles" and "set_1d-model_profiles" are not allowed simultaneously | Set initializing_actions = 'set_constant_profiles' if the 3d-model shall be initialized with constant profiles which can be set with appropriate parameters in the &inipar-Namelist. Set initializing_actions = 'set_1d-model_profiles' if the 3d-model shall be initialized with profiles of the (stationary) solution of the 1d-model. |
| PA0032 ¶ | initializing_actions = "set_constant_profiles" and "by_user" are not allowed simultaneously | Set initializing_actions = 'set_constant_profiles' if the 3d-model shall be initialized with constant profiles which can be set with appropriate parameters in the &inipar-Namelist. Set initializing_actions = 'by_user' if the 3d-model shall be initialized with profiles which can be generated by the user in routine user_init_3d_model of the user-interface. |
| PA0033 ¶ | initializing_actions = "by_user" and "set_1d-model_profiles" are not allowed simultaneously | Set initializing_actions = 'by_user' if the 3d-model shall be initialized with profiles which can be generated by the user in routine user_init_3d_model of the user-interface. Set initializing_actions = 'set_1d-model_profiles' if the 3d-model shall be initialized with profiles of the (stationary) solution of the 1d-model. |
| PA0034 ¶ | cloud_physics = ... is not allowed with humidity = ... | Theres |
| PA0035 ¶ | precipitation = ... is not allowed with cloud_physics = ... | Theres |
| PA0036 ¶ | humidity = .TRUE. and sloping_surface = .TRUE. are not allowed simultaneously | If alpha_surface is set to a non-zero value in order to incline the model domain, the parameter sloping_surface is internally set .TRUE.. I.e., in the case of alpha_surface /= 0.0 the simultaneous use of humidity = .TRUE. is not allowed. |
| PA0037 ¶ | error number free to use | |
| PA0038 ¶ | humidity = .TRUE. and passive_scalar = .TRUE. is not allowed simultaneously' | In PALM, one additional prognostic equation is available to calculate the transport of a scalar quantity. This quantity can either be the humidity or a passive scalar. |
| PA0039 ¶ | error number free to use | |
| PA0040 ¶ | error number free to use | |
| PA0041 ¶ | plant_canopy = .TRUE. requires a non-zero drag coefficient, given value is drag_coefficient = 0.0 | If plant_canopy = .TRUE. a non-zero drag_coefficient must be set to account for the effect of the plant canopy on the flow. The canopy model in PALM adds an additional source/sink term to the momentum equations, the prognostic equation for temperature and the subgrid scale turbulent kinetic energy. In case of humidity = .TRUE. or passive_scalar = .TRUE. an additional source/sink term is added to the prognostic equation of the respective scalar. If drag_coefficient = 0.0, the additional terms equal zero and there is no effect on the flow. |
| PA0042 ¶ | use_top_fluxes must be .TRUE. in ocean version | |
| PA0043 ¶ | ABS( alpha_surface = ... ) must be< 90.0 | Micha |
| PA0044 ¶ | dt = ... <= 0.0 | The parameter dt (time step for the 3d-model) must not be set to a value <= 0.0. |
| PA0045 ¶ | cfl_factor = ... out of range 0.0 < cfl_factor <= 1.0 is required | Matthias |
| PA0046 ¶ | baroclinicity (ug) not allowed simultaneously with galilei transformation | Matthias |
| PA0047 ¶ | baroclinicity (vg) not allowed simultaneously with galilei transformation | Matthias |
| PA0048 ¶ | variable translation speed used for galilei-transformation, which may cause instabilities in stably stratified regions | |
| PA0049 ¶ | unknown boundary condition: bc_lr = "..." | Micha |
| PA0050 ¶ | unknown boundary condition: bc_ns = "..." | Micha |
| PA0051 ¶ | non-cyclic lateral boundaries do not allow psolver = "..." | Using non-cyclic lateral boundary conditions requires the multi-grid method for solving the Poisson equation for the perturbation pressure. You can choose this method by setting the parameter psolver = 'multigrid' . |
| PA0052 ¶ | non-cyclic lateral boundaries do not allow momentum_advec = "..." | Using non-cyclic lateral boundary conditions requires either the scheme of Wicker and Skamarock or the scheme of Piacsek and Williams for the advection of momentum. You can choose one of these methods by setting the parameter momentum_advec. |
| PA0053 ¶ | non-cyclic lateral boundaries do not allow scalar_advec = "..." | Using non-cyclic lateral boundary conditions requires either the scheme of Wicker and Skamarock or the scheme of Piacsek and Williams for the advection of the scalar quantities. You can choose one of these methods by setting the parameter scalar_advec. |
| PA0054 ¶ | non-cyclic lateral boundaries do not allow galilei_transformation = .T. | Jens |
| PA0055 ¶ | error number free to use | |
| PA0056 ¶ | error number free to use | |
| PA0057 ¶ | boundary condition bc_e_b changed to "..." | |
| PA0058 ¶ | unknown boundary condition: bc_e_b = "..." | Micha |
| PA0059 ¶ | unknown boundary condition: bc_p_b = "..." | Micha |
| PA0060 ¶ | boundary condition: bc_p_b = "..." not allowed with prandtl_layer = .FALSE. | |
| PA0061 ¶ | unknown boundary condition: bc_p_t = "..." | Micha |
| PA0062 ¶ | unknown boundary condition: bc_pt_b = "..." | Micha |
| PA0063 ¶ | unknown boundary condition: bc_pt_t = "..." | Micha |
| PA0064 ¶ | both, top_momentumflux_u AND top_momentumflux_v must be set | Jens |
| PA0065 ¶ | boundary_condition: bc_pt_b = "..." is not allowed with constant_heatflux = .TRUE. | |
| PA0066 ¶ | constant_heatflux = .TRUE. is not allowed with pt_surface_initial_change (/=0) = ... | |
| PA0067 ¶ | boundary_condition: bc_pt_t = "..." is not allowed with constant_top_heatflux = .TRUE. | |
| PA0068 ¶ | unknown boundary condition: bc_sa_t = "..." | |
| PA0069 ¶ | boundary condition: bc_sa_t = "..." requires to set top_salinityflux | Jens |
| PA0070 ¶ | boundary condition: bc_sa_t = "..." is not allowed with constant_top_salinityflux = .TRUE.' | Jens |
| PA0071 ¶ | unknown boundary condition: bc_..._b ="..." | |
| PA0072 ¶ | unknown boundary condition: bc_..._t ="..." | |
| PA0073 ¶ | boundary condition: bc_..._b = "..." is not allowed with prescribed surface flux | |
| PA0074 ¶ | a prescribed surface flux is not allowed with ..._surface_initial_change (/=0) = ... | |
| PA0075 ¶ | boundary condition: bc_uv_b = "..." is not allowed with prandtl_layer = .TRUE. | |
| PA0076 ¶ | unknown boundary condition: bc_uv_b = "..." | |
| PA0077 ¶ | unknown boundary condition: bc_uv_t = "..." | |
| PA0078 ¶ | rayleigh_damping_factor = ... out of range [0.0,1.0] | |
| PA0079 ¶ | rayleigh_damping_height = ... out of range [0.0, ...] | |
| PA0080 ¶ | error number free to use | |
| PA0081 ¶ | error number free to use | |
| PA0082 ¶ | number of statistic_regions = ... but only 10 regions are allowed | |
| PA0083 ¶ | normalizing_region = ... must be >= 0 and <= ... (value of statistic_regions) | |
| PA0084 ¶ | dt_sort_particles is reset to 0.0 because of cloud_droplets = .TRUE. | |
| PA0085 ¶ | averaging_interval = ... must be <= dt_data_output = ... | |
| PA0086 ¶ | averaging_interval_pr = ... must be <= dt_dopr = ... | |
| PA0087 ¶ | averaging_interval_sp = ... must be <= dt_dosp = ... | |
| PA0088 ¶ | dt_averaging_input = ... must be <= averaging_interval = ... | |
| PA0089 ¶ | dt_averaging_input_pr = ... must be <= averaging_interval_pr = ... | |
| PA0090 ¶ | precipitation_amount_interval = ... must not be larger than dt_do2d_xy = ... | |
| PA0091 ¶ | data_output_pr = ... is not implemented for ocean = .FALSE. | Jens |
| PA0092 ¶ | data_output_pr = ... is not implemented for humidity = .FALSE. | |
| PA0093 ¶ | data_output_pr = ... is not implemented for passive_scalar = .FALSE. | |
| PA0094 ¶ | data_output_pr = ... is not implemented for cloud_physics = .FALSE. | Theres |
| PA0095 ¶ | data_output_pr = ... is not implemented for cloud_physics = .FALSE. and humidity = .FALSE. | Theres |
| PA0096 ¶ | data_output_pr = ... is not implemented for cloud_physics = .FALSE. or cloud_droplets = .FALSE | Theres |
| PA0097 ¶ | illegal value for data_output_pr or data_output_pr_user = "..." | |
| PA0098 ¶ | illegal value for data_output_pr = "..." | |
| PA0099 ¶ | netCDF: netCDF4 parallel output requested but no cpp-directive __netcdf4_parallel given switch back to netCDF4 non-parallel output | netcdf_data_format > 4 has been set. This requires netCDF4 features including parallel file support, which have to be activated by setting the cpp-switches -D__netcdf4 and -D__netcdf4_parallel in the configuration file (%cpp_options). It also requires a netCDF4 library which supports parallel I/O. Set -I, -L and -l options in compile (%copts) and load (%lopts) options of the configuration file appropriately. Job will be continued with netcdf_data_format = netcdf_data_format - 2. |
| PA0100 ¶ | error number free to use | |
| PA0101 ¶ | error number free to use | |
| PA0102 ¶ | number of output quantities given by data_output and data_output_user exceeds the limit of 100 | |
| PA0103 ¶ | output of "..." requires constant_diffusion = .FALSE. | |
| PA0104 ¶ | output of "..." requires a "particles_par"-NAMELIST in the parameter file (PARIN) | |
| PA0105 ¶ | output of "..." requires humidity = .TRUE. | |
| PA0106 ¶ | output of "..." requires cloud_physics = .TRUE. or cloud_droplets = .TRUE. | |
| PA0107 ¶ | output of "..." requires cloud_droplets = .TRUE. | The quantities ql_c, ql_v, ql_vp are only calculated in case of cloud_droplets = .TRUE.. |
| PA0108 ¶ | output of "..." requires cloud_physics = .TRUE. | For the output of the water vapor content qv (specific humidity) set cloud_physics = .TRUE. to switch on the condensation scheme. Also, humidity = .TRUE. must be set. If you want to simulate the humidity without condensation/evaporation processes, use only humidity = .TRUE. and choose output quantity q for output of the specific humidity. |
| PA0109 ¶ | output of "..." requires ocean = .TRUE. | The quantities rho and sa are only calculated in case of ocean = .TRUE.. |
| PA0110 ¶ | output of "..." requires passive_scalar = .TRUE. | The output of the passive scalar s is only available if the prognostic equation for a passive scalar is solved by setting passive_scalar = .TRUE.. |
| PA0111 ¶ | illegal value for data_output: "..." only 2d-horizontal cross sections are allowed for this value | |
| PA0112 ¶ | output of "..." requires precipitation = .TRUE. | |
| PA0113 ¶ | temporal averaging of precipitation amount "..." is not possible | |
| PA0114 ¶ | illegal value for data_output or data_output_user = "..." | |
| PA0115 ¶ | illegal value for data_output = "..." | |
| PA0117 ¶ | do3d_compress = .TRUE. not allowed on host "..." | |
| PA0118 ¶ | illegal precision: do3d_comp_prec ( ...) = "..." | |
| PA0119 ¶ | unknown variable "..." given for do3d_comp_prec(...) | |
| PA0120 ¶ | unknown value for data_output_format "..." | |
| PA0121 ¶ | km_constant = ... < 0.0 | |
| PA0122 ¶ | prandtl_number = ... < 0.0 | |
| PA0123 ¶ | prandtl_layer is not allowed with fixed value of km | |
PA0124 ¶ | pt_damping_width out of range |
In case of non-cyclic lateral boundary conditions, the parameter pt_damping_width should not be lesser than zero or greater than the model domain length in the respective direction. 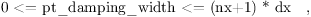 and in case of bc_ns /= 'cyclic': 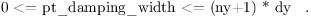 (nx+1), (ny+1) are the total number of grid points and dx, dy the grid lengths in the x-, y-direction, respectively. |
| PA0125 ¶ | rif_min = ... must be less than rif_max = ... | |
| PA0126 ¶ | disturbance_level_b = ... must be >= ... (zu(3)) | |
| PA0127 ¶ | disturbance_level_b = ... must be <= ... (zu(nzt-2)) | |
| PA0128 ¶ | disturbance_level_t = ... must be <= ... (zu(nzt-2)) | |
| PA0129 ¶ | disturbance_level_t = ... must be >= disturbance_level_b = ... | |
| PA0130 ¶ | disturbance_level_ind_t =... must be >= disturbance_level_b = ... | |
| PA0131 ¶ | inflow_disturbance_begin out of range | The parameter inflow_disturbance_begin gives the number of grid points (counted horizontally from the inflow) from which on perturbations are imposed on the horizontal velocity field. This value must not be smaller than zero or larger than nx (in case of non-cyclic lateral boundary conditions along the x-direction) or ny (in case of non-cyclic lateral boundary conditions along the y-direction). |
| PA0132 ¶ | inflow_disturbance_end out of range | The parameter inflow_disturbance_end gives the number of grid points (counted horizontally from the inflow) up to which perturbations are imposed on the horizontal velocity field. This value must not be smaller than zero or larger than nx (in case of non-cyclic lateral boundary conditions along the x-direction) or ny (in case of non-cyclic lateral boundary conditions along the y-direction). |
| PA0133 ¶ | turbulent_inflow = .T. requires a Dirichlet condition at the inflow boundary | Matthias |
| PA0134 ¶ | illegal value for recycling_width: ... | Jens |
| PA0135 ¶ | unknown random generator: random_generator = "..." | |
| PA0136 ¶ | damp_level_1d = ... must be > 0.0 and < ... (zu(nzt+1)) | |
| PA0137 ¶ | mixing_length_1d = "..." is unknown | |
| PA0138 ¶ | dissipation_1d = "..." is unknown | |
| PA0139 ¶ | termination_time_needed = ... must be > 30.0 on host ... | |
| PA0140 ¶ | termination_time_needed = ... should be >= 300.0 on host "..." | |
| PA0141 ¶ | coupling requires PALM to be called with "mrun -K parallel" | |
| PA0142 ¶ | absolute temperature < 0.0 at zu(...) = ... | Within the PALM code (in init_cloud_physics), the hydrostatic pressure profile is calculated for a neutrally stratified temperature profile t(z) = t_surface-g/c_p*z(k). If the height of the model domain is too large, the absolute temperature will become negative, which is not allowed. This error usually appears when both grid stretching is switched on and too many grid points along z are used. Try to reduce the number of grid points (nz), switch on the grid stretching at larger heights (see dz_stretch_level) or reduce the grid stretching factor (dz_stretch_factor). |
| PA0143 ¶ | k=... j=... i=... ql_c=... part(...)%wf=... delta_r=... | |
| PA0144 ¶ | #1 k=... j=... i=... e_s=... e_a=... t_int=... d_radius=... delta_r=... particle_radius=... | Theres |
| PA0145 ¶ | collision_efficiency out of range: ... | Theres |
| PA0146 ¶ | maximum_number_of_particles needs to be increased but this is not allowed with netcdf_data_format < 3 | netCDF output is switched on (data_output_format = 'netcdf' ) and a netCDF3 format is used (netcdf_data_format < 3). If output of particle data is switched on (see dt_write_particle_data), the size of the particle array, given on each processor by maximum_number_of_particles, is not allowed to increase during the run, when the netCDF3 data format is used, because netCDF3 allows only one unlimited dimension, which is the time dimension. Anyhow, PALM tries to increase the size of the particle array, if the number of particles in a subdomain becomes larger than the size of this array. This may happen because of a flow convergence or because new sets of particles are released (see dt_prel). This problem can be avoided by using the netCDF4 data format (netcdf_data_format > 2), which allows more than one unlimited dimension. Alternatively, maximum_number_of_particles can be given a sufficiently large value. |
| PA0147 ¶ | maximum_number_of_tails needs to be increased but this is not allowed with netcdf_data_format < 3 | netCDF output is switched on (data_output_format = 'netcdf' ) and a netCDF3 format is used (netcdf_data_format < 3). If output of particle data is switched on (see dt_write_particle_data), the size of the particle array, given on each processor by maximum_number_of_tails, is not allowed to increase during the run, when the netCDF3 data format is used, because netCDF3 allows only one unlimited dimension, which is the time dimension. Anyhow, PALM tries to increase the size of the particle array, if the number of particles in a subdomain becomes larger than the size of this array. This may happen because of a flow convergence or because new sets of particles are released (see dt_prel). This problem can be avoided by using the netCDF4 data format (netcdf_data_format > 2), which allows more than one unlimited dimension. Alternatively, maximum_number_of_particles can be given a sufficiently large value. |
| PA0148 ¶ | particle too fast. n = ... | |
| PA0149 ¶ | particle out of range: i=... j=... k=... nxl=... nxr=... nys=... nyn=... nzb=... nzt=... | |
| PA0150 ¶ | Both dp_external and conserve_volume_flow are .TRUE. but one of them must be .FALSE. | |
| PA0151 ¶ | dp_level_b = ... is out of range | |
| PA0152 ¶ | dp_external is .TRUE. but dpdxy is zero, i.e. the external pressure gradient will not be applied | |
| PA0153 ¶ | dpdxy is nonzero but dp_external is .FALSE., i.e. the external pressure gradient will not be applied | |
| PA0154 ¶ | unknown conserve_volume_flow_mode: ... | |
| PA0155 ¶ | noncyclic boundary conditions require conserve_volume_flow_mode = "inflow_profile" | Micha |
| PA0156 ¶ | noncyclic boundary conditions require conserve_volume_flow_mode = "inflow_profile" | Micha |
| PA0157 ¶ | nonzero bulk velocity requires conserve_volume_flow = .T. and conserve_volume_flow_mode = "bulk_velocity" | |
| PA0158 ¶ | no vertical boundary condition for variable "..." | |
| PA0159 ¶ | no term for component "..." | |
| PA0160 ¶ | non-cyclic lateral boundaries along x do not allow calculation of spectra along x | Matthias |
| PA0161 ¶ | sorry, calculation of spectra in non parallel mode is still not realized | Matthias |
| PA0162 ¶ | non-cyclic lateral boundaries along y do not allow calculation of spectra along y | Matthias |
| PA0163 ¶ | run will be terminated because it is running out of job cpu limit remaining time: ... s termination time needed: ... s | In case that restart_time has not been set manually, PALM determines internally when a new restart run has to be carried out depending on the setting of termination_time_needed. Within the remaining time PALM will, for example, copy binary files for restart runs, archive or transfer result data. Have a look at Initialization and restart runs for further information. |
| PA0164 ¶ | run will be terminated due to user settings of restart_time / dt_restart new restart time is: ... s | A restart run is established by PALM due to the setting of restart_time and/or dt_restart and the new restart time is calculated. Have a look at Initialization and restart runs for further information. |
| PA0165 ¶ | re-open of unit 14 is not verified. Please check results! | |
| PA0166 ¶ | re-opening of file-id ... is not allowed | |
| PA0167 ¶ | opening file-id ... not allowed for PE ... | |
| PA0168 ¶ | opening file-id ... is not allowed since it is used otherwise | |
| PA0169 ¶ | no filename for AVS-data-file found in MRUN-config-file filename in FLD-file set to "unknown" | |
| PA0170 ¶ | no path for batch_scp on host "..." | |
| PA0171 ¶ | netCDF: netCDF4 format requested but no cpp-directive __netcdf4 given switch back to 64-bit offset format | netcdf_data_format > 2 has been set. This requires netCDF4 features, which have to be activated by setting the cpp-directive __netcdf4 in the configuration file (%cpp_options). It also requires a netCDF4 library. Set -I, -L and -l options in compile (%copts) and load (%lopts) options of the configuration file appropriately. |
| PA0172 ¶ | no OPEN-statement for file-id ... | |
| PA0173 ¶ | wrong component: ... | |
| PA0174 ¶ | wrong argument expected: ... given: | Internal code error. Please inform the PALM developers. |
| PA0175 ¶ | no time measurement defined on this host | |
| PA0176 ¶ | negative time interval occured PE ... L=PAUSE "..." new=... last=... | |
| PA0177 ¶ | negative time interval occured PE ... L=STOP "..." new=... last=... isum=... | |
| PA0178 ¶ | negative time interval occured PE ... L=STOP "..." sum=... mtime=... | |
| PA0179 ¶ | unknown modus of time measurement: ... | |
| PA0180 ¶ | unknown cross-section: ... | |
| PA0181 ¶ | no output provided for: ... | |
| PA0182 ¶ | if humidity/passive_scalar = FALSE output of ... is not provided | |
| PA0183 ¶ | if cloud_physics = FALSE output of ... is not provided | Theres |
| PA0184 ¶ | if cloud_physics = FALSE output of ... is not provided | Theres |
| PA0185 ¶ | data_output_profiles: normalizing cross ... is not possible since one of the normalizing factors is zero! cross_normx_factor(...,...) = ... cross_normy_factor(...,...) = ... | |
| PA0186 ¶ | no spectra data available | |
| PA0187 ¶ | fft method "..." currently does not work on NEC | Siggi |
| PA0188 ¶ | no system-specific fft-call available | Siggi |
| PA0189 ¶ | fft method "..." not available | Siggi |
| PA0190 ¶ | flow_statistics is called two times within one timestep | |
| PA0191 ¶ | unknown action(s): ... | |
| PA0192 ¶ | timestep has exceeded the lower limit dt_1d = ... s simulation stopped! | |
| PA0193 ¶ | unknown initializing problem | |
| PA0194 ¶ | number of time series quantities exceeds its maximum of dots_max = ... Please increase dots_max in modules.f90. | In case that the output of user defined timeseries is done and the number of the user defined timeseries exceeds the maximum number, the maximum number dots_max, which is currently set to 100, has to be increased. Please increase dots_max in modules.f90 according to your needs. |
| PA0195 ¶ | dvrp_username is undefined | |
| PA0196 ¶ | dvrp_output="..." not allowed | |
| PA0197 ¶ | dvrp_file="..." not allowed | |
| PA0198 ¶ | mode_dvrp="..." not allowed | |
| PA0199 ¶ | split of communicator not realized with MPI1 coupling atmosphere-ocean' | |
| PA0200 ¶ | missing dz | |
| PA0201 ¶ | dz=... <= 0.0 | |
| PA0202 ¶ | grid anisotropy exceeds threshold given by only local horizontal reduction of near_wall mixing length l_wall starting from height level k = ... . | |
| PA0203 ¶ | inconsistent building parameters: bxl=... bxr=... bys=... byn=... nx=... ny=... | |
| PA0204 ¶ | no street canyon width given | |
| PA0205 ¶ | inconsistent canyon parameters: cxl=... cxr=... cwx=... ch=... nx=... ny=... | |
| PA0206 ¶ | inconsistent canyon parameters:... cys=... cyn=... cwy=... ch=... nx=... ny=... | |
| PA0207 ¶ | inconsistent canyon parameters: street canyon can only be oriented either in x- or in y-direction | |
| PA0208 ¶ | file TOPOGRAPHY_DATA does not exist | |
| PA0209 ¶ | errors in file TOPOGRAPHY_DATA | Possible reasons for this error include: 1. File contains a header. Solution: The header of the ESRI Ascii Grid format (usually 6 lines) must be removed. 2. File does not contain enough data. a) The number of columns (ncols) is less than nx+1. b) The number of rows (nrows) is less than ny+1. Solution: Make sure that ncols matches nx+1 and that nrows matches ny+1. |
| PA0210 ¶ | nzb_local values are outside the model domain MINVAL( nzb_local ) = ... MAXVAL( nzb_local ) = ... | |
| PA0211 ¶ | nzb_local does not fulfill cyclic boundary condition in x-direction | |
| PA0212 ¶ | nzb_local does not fulfill cyclic boundary condition in y-direction | |
| PA0213 ¶ | max_number_of_particle_groups =... number_of_particle_groups reset to ... | |
| PA0214 ¶ | version mismatch concerning data from prior run version on file = ... version in program = ... | |
| PA0215 ¶ | particle group !# ... has a density ratio /= 0 but radius = 0 | |
| PA0216 ¶ | number of initial particles (...) exceeds maximum_number_of_particles (...) on PE ... | Theres |
| PA0217 ¶ | unknown boundary condition bc_par_b = "..." | |
| PA0218 ¶ | unknown boundary condition bc_par_t = "..." | |
| PA0219 ¶ | unknown boundary condition bc_par_lr = "..." | |
| PA0219 ¶ | unknown boundary condition bc_par_ns = "..." | |
| PA0221 ¶ | number of PEs of the prescribed topology (...) does not match the number of PEs available to the job (...) | In case that the number of PEs along the x- and y-direction of the virtual processor grid is set via npex and npey, the product npey*npex has to match exactly the total number of processors which is assigned by the mrun-option -X. |
| PA0222 ¶ | if the processor topology is prescribed by the user, both values of "npex" and "npey" must be given in the NAMELIST-parameter file | In case that only one number of processors along x- or y- direction npex or npey is set in the &d3par-Namelist, the remaining, so far not assigned, number of processors has to be set, too. |
| PA0223 ¶ | psolver = "poisfft_hybrid" can only be used in case of a 1d-decomposition along x | Siggi |
| PA0224 ¶ | no matching grid for transpositions found | |
| PA0225 ¶ | x-direction: gridpoint number (...) is not an integral divisor of the number of processors (...) | The division of the number of grid points along the x-direction given by nx+1 by the number of processors along the x-direction has a rest and is therefore not an integral divisor of the number of processors. To solve this problem you must ensure that the aforementioned division is without rest for example by setting the number of PEs in y-direction manually (see npex) or by changing nx. It might also be the case that you did not think about that nx+1 is the total number of gridpoints along the x-direction instead of nx. |
| PA0226 ¶ | x-direction: nx does not match the requirements given by the number of PEs used please use nx = ... instead of nx =... | |
| PA0227 ¶ | y-direction: gridpoint number (...) is not an integral divisor of the number of processors (...) | The division of the number of grid points along the y-direction given by ny+1 by the number of processors along the y-direction has a rest and is therefore not an integral divisor of the number of processors. To solve this problem you must ensure that the aforementioned division is without rest for example by setting the number of PEs in y-direction manually (see npey) or by changing ny. It might also be the case that you did not think about that ny+1 is the total number of gridpoints along the y-direction instead of ny. |
| PA0228 ¶ | y-direction: ny does not match the requirements given by the number of PEs used please use ny = ... instead of ny =... | |
| PA0229 ¶ | error number free to use | |
| PA0230 ¶ | transposition z --> x: nz=... is not an integral divisor of pdims(1)= ... | Due to restrictions in data transposition, the division of nz by the number of processors in x-direction has to be without rest. Please ensure this by changing nz or setting the number of gridpoints in x-direction (pdims(1)) manually by npex. |
| PA0231 ¶ | transposition x --> y: nx+1=... is not an integral divisor of pdims(2)= ... | Due to restrictions in data transposition, the division of nx+1 by the number of processors in y-direction has to be without rest. Please ensure this by changing nx or setting the number of gridpoints in y-direction (pdims(2)) manually by npex. |
| PA0232 ¶ | transposition y --> z: ny+1=... is not an integral divisor of pdims(1)=... | Due to restrictions in data transposition, the division of ny+1 by the number of processors in x-direction has to be without rest. Please ensure this by changing ny or setting the number of gridpoints in x-direction (pdims(1)) manually by npex. |
| PA0233 ¶ | transposition x --> y: ny+1=... is not an integral divisor of pdims(1)=... | Due to restrictions in data transposition, the division of ny+1 by the number of processors in x-direction has to be without rest. Please ensure this by changing ny or setting the number of gridpoints in x-direction (pdims(1)) manually by npex. |
| PA0234 ¶ | direct transposition z --> y (needed for spectra): nz=... is not an integral divisor of pdims(2)=... | |
| PA0235 ¶ | mg_switch_to_pe0_level out of range and reset to default (=0) | |
| PA0236 ¶ | grid coarsening on subdomain level cannot be performed | The multigrid pressure solver halfs the number of grid points in each direction to get coarser grid levels. This is done for the subdomains on each PE until one of the directions cannot be divided by 2 without rest. If this is the case, the coarsest grid level of each subdomain are gathered on each PE for further coarsening. Before gathering, the coarsening must be done for the subdomain at least one time. You get this error messages for example with the following combination: nx = 60, ny = 60, nz = 60, npex = 4, npey = 2. With this combination, the subdomain on each PE has the size: nx = 15, ny = 30, nz = 60. The number of grid points in the x-direction of the subdomain cannot be divided by 2 and no coarsening is possible for the subdomains in this example. To avoid this error ensure that the number of grid points in each direction can be divided by 2 without rest on the subdomain of an PE at least one time. Therefor vary the number of grid points for the direction which does not fit by changing nx , ny or nz or adjust the number of processors by modifying the parameters npex or npey in order to get other subdomain sizes. In the example given above, you can avoid the error by changing nx or npex. For example setting nx = 64 leads to a subdomain of nx = 16, ny = 30, nz = 60. All numbers are now divisible by 2 without rest and the first coarser grid on the subdomain is then: nx = 8, ny = 15, nz = 30. |
| PA0237 ¶ | multigrid gather/scatter impossible in non parallel mode | Siggi |
| PA0238 ¶ | more than 10 multigrid levels | Siggi |
| PA0239 ¶ | The value for "topography_grid_convention" is not set. Its default value is only valid for "topography" = "single_building", "single_street_canyon" or "read_from_file" Choose "cell_edge" or "cell_center". | |
| PA0240 ¶ | The value for "topography_grid_convention" is not recognized. Choose "cell_edge" or "cell_center". | |
| PA0241 ¶ | netcdf_precision must contain a "_" netcdf_precision(...)="..." | |
| PA0242 ¶ | illegal netcdf precision: netcdf_precision( ...)="..." | |
| PA0243 ¶ | unknown variable in inipar assignment: netcdf_precision(...)="..." | |
| PA0244 ¶ | no grid defined for variable ... | |
| PA0245 ¶ | netCDF file for volume data ... from previous run found, but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0246 ¶ | netCDF file for volume data ... from previous run found, but this file cannot be extended due to mismatch in number of vertical grid points (nz_do3d). New file is created 47stead. | |
| PA0247 ¶ | netCDF file for volume data ... from previous run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0248 ¶ | netCDF file for volume data ... from previous run found. This file will be extended. | |
| PA0249 ¶ | netCDF file for cross-sections ... from previous run found, but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0250 ¶ | netCDF file for cross-sections ... from previous run found, but this file cannot be extended due to mismatch in number of cross sections. New file is created instead. | |
| PA0251 ¶ | netCDF file for cross-sections ... from previuos run found but this file cannot be extended due to mismatch in cross section levels. New file is created instead. | |
| PA0252 ¶ | netCDF file for cross sections ... from previous run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0253 ¶ | netCDF file for cross-sections ... from previous run found. This file will be extended. | |
| PA0254 ¶ | netCDF file for vertical profiles from previous run found, but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0255 ¶ | netCDF file for vertical profiles from previous run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0256 ¶ | netCDF file for vertical profiles from previous run found. This file will be extended. | |
| PA0257 ¶ | netCDF file for vertical profiles from previous run found. This file will be extended. | |
| PA0258 ¶ | netCDF file for time series from previous run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0259 ¶ | netCDF file for time series from previous run found. This file will be extended. | |
| PA0260 ¶ | netCDF file for spectra from previuos run found, but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0261 ¶ | netCDF file for spectra from previuos run found, but this file cannot be extended due to mismatch in number of vertical levels. New file is created instead. | |
| PA0262 ¶ | netCDF file for spectra from previuos run found, but this file cannot be extended due to mismatch in heights of vertical levels. New file is created instead. | |
| PA0263 ¶ | netCDF file for spectra from previuos run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0264 ¶ | netCDF file for spectra from previous run found. This file will be extended. | |
| PA0265 ¶ | netCDF file for particles from previuos run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0266 ¶ | netCDF file for particles from previous run found. This file will be extended. | |
| PA0267 ¶ | netCDF file for particle time series from previuos run found, but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0268 ¶ | netCDF file for particle time series from previuos run found, but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0269 ¶ | netCDF file for particle time series from previous run found. This file will be extended. | |
| PA0270 ¶ | mode "..." not supported | |
| PA0271 ¶ | errors in $inipar or $inipar-namelist found (CRAY-machines only) | One possible explanation: An old, meanwhile modified variable has been used in $inipar (e.g. ws_vertical_gradient (old) instead of subs_vertical_gradient (new)) |
| PA0272 ¶ | no $inipar-namelist found | Farah |
| PA0273 ¶ | no value or wrong value given for nx: nx=... | |
| PA0274 ¶ | no value or wrong value given for ny: ny=... | |
| PA0275 ¶ | no value or wrong value given for nz: nz=... | |
| PA0276 ¶ | local file ENVPAR not found some variables for steering may not be properly set | |
| PA0277 ¶ | errors in local file ENVPAR some variables for steering may not be properly set | |
| PA0278 ¶ | no envpar-NAMELIST found in local file ENVPAR some variables for steering may not be properly set | |
| PA0279 ¶ | wrong component: ... | |
| PA0280 ¶ | Number of OpenMP threads = ... | |
| PA0281 ¶ | running optimized multinode version switch_per_lpar = ... tasks_per_lpar = ... tasks_per_logical_node = ... | |
| PA0282 ¶ | parallel environment (MPI) required | |
| PA0283 ¶ | no sufficient convergence within 1000 cycles | Siggi |
| PA0284 ¶ | data from subdomain of previous run mapped more than 1000 times | |
| PA0285 ¶ | number of PEs or virtual PE-grid changed in restart run PE ... will read from files ... | |
| PA0286 ¶ | version mismatch concerning data from prior run version on file = "..." version in program = "..." | |
| PA0287 ¶ | problem with index bound nxl on restart file "..." nxl = ... but it should be = ... from the index bound information array | |
| PA0288 ¶ | problem with index bound nxr on restart file "..." nxr = ... but it should be = ... from the index bound information array | |
| PA0289 ¶ | problem with index bound nys on restart file "..." nys = ... but it should be = ... from the index bound information array | |
| PA0290 ¶ | problem with index bound nyn on restart file "..." nyn = ... but it should be = ... from the index bound information array | |
| PA0201 ¶ | mismatch between actual data and data from prior run on PE ... nzb on file = ... nzb = ... | |
| PA0292 ¶ | mismatch between actual data and data from prior run on PE ... nzt on file = ... nzt = ... | |
| PA0293 ¶ | read_3d_binary: spectrum_x on restart file ignored because total numbers of grid points (nx) do not match | |
| PA0294 ¶ | read_3d_binary: spectrum_y on restart file ignored because total numbers of grid points (ny) do not match | |
| PA0295 ¶ | unknown field named "..." found in data from prior run on PE ... | |
| PA0296 ¶ | version mismatch concerning control variables version on file = "..." version on program = "..." | |
| PA0297 ¶ | numprocs not found in data from prior run on PE ... | |
| PA0298 ¶ | hor_index_bounds not found in data from prior run on PE ... | |
| PA0299 ¶ | nz not found in data from prior run on PE ... | |
| PA0300 ¶ | max_pr_user not found in data from prior run on PE ... | |
| PA0301 ¶ | statistic_regions not found in data from prior run on PE ... | |
| PA0302 ¶ | unknown variable named "..." found in data from prior run on PE ... | |
| PA0303 ¶ | nz not found in restart data file | |
| PA0304 ¶ | mismatch concerning number of gridpoints along z nz on file = "..." nz from run = "..." | |
| PA0305 ¶ | max_pr_user not found in restart data file | |
| PA0306 ¶ | number of user profiles on restart data file differs from the current run max_pr_user on file = "..." max_pr_user from run = "..." | |
| PA0307 ¶ | statistic_regions not found in restart data file | |
| PA0308 ¶ | statistic regions on restart data file differ from the current run statistic regions on file = "..." statistic regions from run = "..." statistic data may be lost! | |
| PA0309 ¶ | inflow profiles not temporally averaged. Averaging will be done now using ... samples. | |
| PA0310 ¶ | remote model "..." terminated with terminate_coupled_remote = ... local model "..." has terminate_coupled = ... | |
| PA0311 ¶ | number of gridpoints along x or/and y contain illegal factors only factors 8,6,5,4,3,2 are allowed | In case that fft_method='temperton-algorithm', the number of horizontal grid points (nx+1, ny+1) must be composed of prime factors 2,3 and 5. Higher prime factors such as 7 are not allowed. Please ensure that these restrictions are fulfilled or use 'singleton-algorithm' as fft_method. |
| PA0312 ¶ | Time step has reached minimum limit. dt = ... s Simulation is terminated. old_dt = ... s dt_u = ... s dt_v = ... s dt_w = ... s dt_diff = ... s u_max = ... m/s k=... j=... i=... v_max = ... m/s k=... j=... i=...\w_max = ... m/s k=... j=... i=... | Matthias |
| PA0313 ¶ | illegal value for parameter particle_color: ... | Farah |
| PA0314 ¶ | illegal value for parameter particle_dvrpsize: ... | Farah |
| PA0315 ¶ | color_interval(2) <= color_interval(1) | |
| PA0316 ¶ | dvrpsize_interval(2) <= dvrpsize_interval(1) | |
| PA0317 ¶ | ocean = .F. does not allow coupling_char = ... set by mrun-option "-y" | The mrun option "-y" must not be used for atmosphere runs. The option is only required for ocean precursor runs, which are followed by atmosphere-ocean coupled restart runs. |
| PA0318 ¶ | inflow_damping_height must be explicitly specified because the inversion height calculated by the prerun is zero | |
| PA0319 ¶ | section_xy must be <= nz + 1 = ... | |
| PA0320 ¶ | section_xz must be <= ny + 1 = ... | |
| PA0321 ¶ | section_yz must be <= nx + 1 = ... | |
| PA0322 ¶ | output of q*w*(0) requires humidity=.TRUE. | Farah |
| PA0323 ¶ | output of averaged quantity "..." requires to set a non-zero averaging interval | Farah |
| PA0324 ¶ | Applying large scale vertical motion is not allowed for ocean runs | The large scale subsidence is only reasonable and implemented for atmospheric runs. The setting of subs_vertical_gradient and subs_vertical_gradient_level is not possible. |
| PA0325 ¶ | illegal value: masks must be >= 0 and <= "..."(=max_masks) | Björn |
| PA0326 ¶ | illegal value: mask_scale_x, mask_scale_y and mask_scale_z must be > 0.0 | |
| PA0327 ¶ | no output available for: "..." | |
| PA0328 ¶ | netCDF file formats 3 (netCDF 4) and 4 (netCDF 4 Classic model) are currently not supported (not yet tested). | |
| PA0329 ¶ | number of output quantitities given by data_output_mask and data_output_mask_user exceeds the limit of 100 | |
| PA0330 ¶ | illegal value for data_output: "..." | |
| PA0331 ¶ | "..." in mask "..." along dimension "..." exceeds "..." = "..." | Björn |
| PA0332 ¶ | mask_loop("...","...",1)="..." and/or mask_loop("...","...",2)="..." exceed "..."*"..."/="..." | Björn |
| PA0333 ¶ | mask_loop("...","...",1)="..." and/or mask_loop("...","...",2)="..." exceed zw(nz)/mask_scale("...")="..." | |
| PA0334 ¶ | mask_loop("...","...",2)="..." exceeds dz_stretch_level="...". Vertical mask locations will not match the desired heights within the stretching region. Recommendation: use mask instead of mask_loop. | Björn |
| PA0335 ¶ | netCDF file for "..." data for mask "..." from previous run found but this file cannot be extended due to variable mismatch. New file is created instead. | |
| PA0336 ¶ | netCDF file for "..." data for mask "..." from previous run found but this file cannot be extended due to mismatch in number of vertical grid points. New file is created instead. | |
| PA0337 ¶ | netCDF file for "..." data for mask "..." from previous run found but this file cannot be extended because the current output time is less or equal than the last output time on this file. New file is created instead. | |
| PA0338 ¶ | netCDF file for "..." data for mask "..." from previous run found. This file will be extended. | |
PA0339 ¶ | coupling mode "...: nx+1 in ocean is not divisible without remainder with nx+1 in atmosphere | For the coupling between atmosphere and ocean, a smaller grid length in the x-direction can be chosen for the ocean, so that the ocean has more grid points in the x-direction then the atmosphere. In this case, the information must be interpolated for the data exchange between atmosphere and ocean. Therefor it must be ensured that each interpolation interval has the same number of gridpoints. This is ensured if the following equation is fulfilled: with
as the number of grid points in the x-direction for the ocean and atmosphere model, respectively. |
PA0340 ¶ | coupling mode "...: ny+1 in ocean is not divisible without remainder with ny+1 in atmosphere | For the coupling between atmosphere and ocean, a smaller grid length in the y-direction can be chosen for the ocean, so that the ocean has more grid points in the y-direction then the atmosphere. In this case, the information must be interpolated for the data exchange between atmosphere and ocean. Therefor it must be ensured that each interpolation interval has the same number of gridpoints. This is ensured if the following equation is fulfilled: with
as the number of grid points in the y-direction for the ocean and atmosphere model, respectively. |
| PA0343 ¶ | initializing_actions = 'initialize_vortex' ist not allowed with conserve_volume_flow = .T. | |
| PA0344 ¶ | psolver must be called at each RK3 substep when 'ws-scheme' is used for momentum_advec. | Matthias |
| PA0345 ¶ | uv_heights(1) must be 0.0 | |
| PA0346 ¶ | u_profile(1) and v_profile(1) must be 0.0 | |
| PA0347 ¶ | maxtry > 40 in Rosenbrock method | Error may appear if curvature and solution effects are included in the calculation of droplet growth by condensation (see curvature_solution effects. It means that the Rosenbrock method, used for integrating the stiff droplet growth equation, was unable to find an internal timestep which gives a sufficiently small truncation error within 40 tries. So far, this problem has not appeared. It might happen if some of the cloud physics parameters, which determine the growth by condensation, are given unrealistic values or if the droplet radius is extremely small. |
| PA0348 ¶ | zero stepsize in Rosenbrock method | Error may appear if curvature and solution effects are included in the calculation of droplet growth by condensation (see curvature_solution effects. It means that within the Rosenbrock method, used for integrating the stiff droplet growth equation, the internal timestep has become zero, so that the integration cannot be performed. So far, this problem has not appeared. It might happen if some of the cloud physics parameters, which determine the growth by condensation, are given unrealistic values or if the droplet radius is extremely small. |
| PA0349 ¶ | use_sgs_for_particles = .TRUE. not allowed with curvature_solution_effects = .TRUE. | When Lagrangian particles are used as cloud droplets, they are only allowed to be transported with the resolved-scale flow. The application of the stochastic subgrid-scale transport model is limited to passively advected particles. Since cloud droplets have inertia, a transport with the SGS model would give wrong results. |
| PA0350 ¶ | unknown collision kernel: collision_kernel = "..." | see package parameter collision_kernel for allowed values |
| PA0351 ¶ | heatflux must not be set for pure neutral flow | Inipar parameter neutral = .TRUE. has been set, but non-zero values for parameters surface_heatflux and / or top_heatflux are given at the same time. This is not allowed for simulating a flow with pure neutral stratification where the temperature equation is switched off. Check your parameter file. |
| PA0352 ¶ | unknown boundary condition: bc_qr_b = "..." | Unknown lower boundary condition for rain water content. Allowed boundary conditions are "dirichlet" or "neumann". |
| PA0353 ¶ | unknown boundary condition: bc_qr_t = "..." | Unknown upper boundary condition for rain water content. Allowed boundary conditions are "dirichlet" or "neumann". |
| PA0354 ¶ | It is not allowed to arrange more than 100 profiles with cross_profiles. Apart from that, all profiles are saved to the netCDF file. | All profiles are written to the netCDF file. Only the first 100 profiles will be plotted by palmplot pr with var=cross_profiles (default). To see all profiles, add the parameter var=all to palmplot pr. |
| PA0355 ¶ | unknown boundary condition: bc_nr_b = "..." | Unknown lower boundary condition for rain drop number density. Allowed boundary conditions are "dirichlet" or "neumann". |
| PA0356 ¶ | unknown boundary condition: bc_nr_t = "..." | Unknown upper boundary condition for rain drop number density. Allowed boundary conditions are "dirichlet" or "neumann". |
| PA0357 ¶ | unknown cloud microphysics scheme cloud_scheme = "..." | Unknown cloud microphysics scheme. Allowed cloud microphysics schemes are "kessler" or "seifert_beheng". |
| PA0358 ¶ | data_output_pr = '...' is not implemented for cloud_scheme /= seifert_beheng | Output of profiles of rain drop number concentration (nr), rain water content (qr) or precipitation rate (prr) is only allowed for cloud_scheme = "seifert_beheng". |
| PA0359 ¶ | output of "..." requires cloud_scheme = seifert_beheng | Output of 3d-data or cross-sections of rain drop number concentration (nr), rain water content (qr) or precipitation rate (prr) is only allowed for cloud_scheme = "seifert_beheng". |
| PA0360 ¶ | 'plant_canopy = .TRUE. requires cloud_scheme /= seifert_beheng' | It is not allowed to use plant_canopy with the cloud_scheme = "seifert_beheng". |
| PA0361 ¶ | data_output_pr = prr is not implemented for precipitation = .FALSE. | Output of precipitation rate (prr) 3d-data or cross-sections requires precipitation = .TRUE. |
| PA0362 ¶ | cloud_scheme = seifert_beheng requires loop_optimization = cache | Currently, the cloud microphysics scheme according to Seifert and Beheng (2006) is only available for loop_optimization = "cache" |
| PA0363 ¶ | cloud_scheme = seifert_beheng requires precipitation = .TRUE. | cloud_scheme = "seifert_beheng" and cloud_scheme = "kessler" are identically if precipitation = .FALSE.. In order to save computational resources, it is not allowed to use cloud_scheme = "seifert_beheng" without precipitation = .TRUE. |
| UI0001 ¶ | unknown location "..." | |
| UI0002 ¶ | location "..." is not allowed to be called with parameters "i" and "j" | |
| UI0003 ¶ | no output possible for: ... | |
| UI0004 ¶ | unknown mode "..." | |
| UI0005 ¶ | topography "..." not available yet | |
| UI0006 ¶ | unknown topography "..." | |
| UI0007 ¶ | canopy_mode "..." not available yet | |
| UI0008 ¶ | unknown canopy_mode "..." | |
| UI0009 ¶ | the number of user-defined profiles given in data_output_pr (...) does not match the one found in the restart file (...) | |
| UI0010 ¶ | Spectra of ... can not be calculated | |
| UI0011 ¶ | Spectra of ... are not defined | |
| UI0012 ¶ | unknown variable named "..." found in data from prior run on PE | |
| NC**** ¶ | **** contains numbers in the range from 1-521 | Internally, PALM calls the netCDF function NF90_STRERROR which returns an error message string provided by netCDF. Hence, no exact assignment between PALM error number and netCDF error string can be given. Most of the netCDF errors should never appear if the default code is used. The following list gives errors which may be caused by wrong steering or errors due to problems in the user interface. |
| NC0079 ¶ | String match to name in use | A netCDF variable name has been used twice. Possible reasons: a variable in the parameter file has been listed twice (e.g. in data_output) or a variable name that has been defined in the user interface is already used by PALM. |
| NC0253 ¶ | Name contains illegal characters | netCDF variable names consist of arbitrary sequences of alphanumeric characters. These names are e.g. assigned by the user in the user-interface to variables dots_label or dopr_label. Due to the netCDF convention, they must not start with a digit. Also, usage of / as special character is not allowed. Older netCDF versions (before 3.6.3) have further restrictions on special characters. Please check if your string settings in the user-interface follow the above rules. |
